Find Edges in Shortest Paths
HardYou are given an undirected weighted graph of n nodes numbered from 0 to n - 1. The graph consists of m edges represented by a 2D array edges, where edges[i] = [ai, bi, wi] indicates that there is an edge between nodes ai and bi with weight wi.
Consider all the shortest paths from node 0 to node n - 1 in the graph. You need to find a boolean array answer where answer[i] is true if the edge edges[i] is part of at least one shortest path. Otherwise, answer[i] is false.
Return the array answer.
Note that the graph may not be connected.
Example 1:
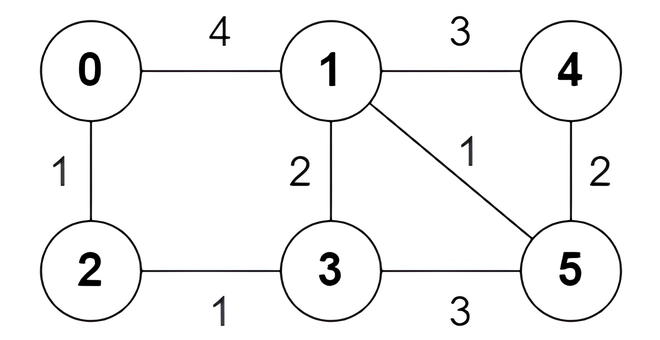
Input: n = 6, edges = [[0,1,4],[0,2,1],[1,3,2],[1,4,3],[1,5,1],[2,3,1],[3,5,3],[4,5,2]]
Output: [true,true,true,false,true,true,true,false]
Explanation:
The following are all the shortest paths between nodes 0 and 5:
- The path
0 -> 1 -> 5: The sum of weights is4 + 1 = 5. - The path
0 -> 2 -> 3 -> 5: The sum of weights is1 + 1 + 3 = 5. - The path
0 -> 2 -> 3 -> 1 -> 5: The sum of weights is1 + 1 + 2 + 1 = 5.
Example 2:
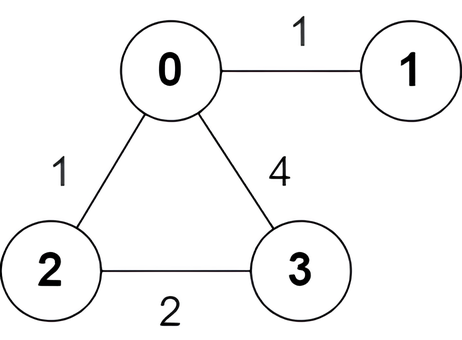
Input: n = 4, edges = [[2,0,1],[0,1,1],[0,3,4],[3,2,2]]
Output: [true,false,false,true]
Explanation:
There is one shortest path between nodes 0 and 3, which is the path 0 -> 2 -> 3 with the sum of weights 1 + 2 = 3.
Constraints:
2 <= n <= 5 * 104m == edges.length1 <= m <= min(5 * 104, n * (n - 1) / 2)0 <= ai, bi < nai != bi1 <= wi <= 105- There are no repeated edges.
Solution
Clarifying Questions
When you get asked this question in a real-life environment, it will often be ambiguous (especially at FAANG). Make sure to ask these questions in that case:
- Are the node IDs represented as integers, and what is the range of possible node IDs (e.g., 0 to n-1 or 1 to n)?
- What should be returned if there is no path between the source and destination nodes?
- Can there be multiple edges between the same two nodes (i.e., parallel edges), and if so, should they be treated as distinct edges for the purpose of identifying edges on shortest paths?
- Are the edge weights guaranteed to be non-negative? If negative weights are allowed, does the graph contain negative cycles?
- Is the graph guaranteed to be connected? If not, are we only concerned with the connected component containing the source and destination nodes?
Brute Force Solution
Approach
To find the edges in the shortest paths between two points, we can try every possible path. We'll build up each path step by step, checking if it's a valid path and calculating its length.
Here's how the algorithm would work step-by-step:
- Start by listing out all possible paths from the starting point to the ending point. Imagine exploring every route possible, one at a time.
- For each path, figure out how long it is. This is done by adding up the distances along each connection in the path.
- Find the shortest distance among all the paths we checked. This is our target shortest distance.
- Now, go back through each path again. Check if the path's length is equal to the shortest distance we found.
- If a path is one of the shortest, look at each connection (edge) used in that path.
- Keep track of all the connections that are part of at least one shortest path. These are the edges we want to find.
- In the end, you will have identified all the connections that are used in at least one of the shortest possible routes between the start and end points.
Code Implementation
def find_edges_in_shortest_paths_brute_force(graph, start_node, end_node):
all_paths = find_all_paths(graph, start_node, end_node)
if not all_paths:
return []
path_distances = {}
for path in all_paths:
distance = calculate_path_distance(graph, path)
path_distances[tuple(path)] = distance
shortest_distance = min(path_distances.values())
shortest_paths = [path for path, distance in path_distances.items() if distance == shortest_distance]
shortest_path_edges = set()
# Iterate through shortest paths to find edges
for path in shortest_paths:
for i in range(len(path) - 1):
shortest_path_edges.add((path[i], path[i+1]))
return list(shortest_path_edges)
def find_all_paths(graph, start_node, end_node, path=None):
if path is None:
path = [start_node]
if start_node == end_node:
return [path]
if start_node not in graph:
return []
paths = []
for neighbor in graph[start_node]:
if neighbor not in path:
new_path = path + [neighbor]
new_paths = find_all_paths(graph, neighbor, end_node, new_path)
paths.extend(new_paths)
return paths
def calculate_path_distance(graph, path):
total_distance = 0
# Sum up the edge weights for this path
for i in range(len(path) - 1):
node_1 = path[i]
node_2 = path[i+1]
if node_2 not in graph[node_1]:
return float('inf')
total_distance += graph[node_1][node_2]
return total_distanceBig(O) Analysis
Optimal Solution
Approach
The problem asks us to identify edges that lie on any shortest path between two given nodes in a graph. The optimal approach involves first finding the shortest path distances and then checking each edge to see if it could be part of a shortest path.
Here's how the algorithm would work step-by-step:
- First, find the shortest distance from the starting node to every other node in the graph. We can use a well-known algorithm like Dijkstra's algorithm or Breadth-First Search for this.
- Next, find the shortest distance from the ending node to every other node in the graph using the same algorithm.
- Now, consider each edge in the graph. For a given edge, check if the sum of the shortest distance from the start node to the edge's source, the weight of that edge, and the shortest distance from the edge's destination to the end node is equal to the overall shortest distance from the start node to the end node. If it is, this edge lies on at least one shortest path.
- Also repeat the check considering the reverse direction of the edge. If the graph is directed, simply consider the direction of the edge.
- Return all the edges that satisfied the condition in the previous step as they are the edges that exist on any shortest path.
Code Implementation
from collections import deque
def find_edges_in_shortest_paths(graph, start_node, end_node):
shortest_distance = find_shortest_distance(graph, start_node, end_node)
if shortest_distance == float('inf'):
return []
edges_in_shortest_paths = set()
queue = deque([(start_node, [start_node])])
while queue:
current_node, path = queue.popleft()
if current_node == end_node and len(path) - 1 == shortest_distance:
# Found a shortest path, add edges to the result
for i in range(len(path) - 1):
edges_in_shortest_paths.add(tuple(sorted((path[i], path[i+1]))))
else:
for neighbor in graph[current_node]:
if len(path) <= shortest_distance:
queue.append((neighbor, path + [neighbor]))
return list(edges_in_shortest_paths)
def find_shortest_distance(graph, start_node, end_node):
distances = {node: float('inf') for node in graph}
distances[start_node] = 0
queue = deque([start_node])
while queue:
current_node = queue.popleft()
for neighbor in graph[current_node]:
new_distance = distances[current_node] + 1
if new_distance < distances[neighbor]:
# Found a shorter path, update distance
distances[neighbor] = new_distance
queue.append(neighbor)
return distances[end_node]
def find_edges_in_shortest_paths_optimized(graph, start_node, end_node):
shortest_distance = find_shortest_distance(graph, start_node, end_node)
if shortest_distance == float('inf'):
return []
edges_in_shortest_paths = set()
queue = deque([(start_node, [start_node])])
while queue:
current_node, current_path = queue.popleft()
if current_node == end_node and len(current_path) - 1 == shortest_distance:
# Add edges only when shortest path is found
for i in range(len(current_path) - 1):
edges_in_shortest_paths.add(tuple(sorted((current_path[i], current_path[i + 1]))))
else:
if len(current_path) -1 < shortest_distance:
for neighbor in graph[current_node]:
# Only consider neighbors to avoid infinite loops
queue.append((neighbor, current_path + [neighbor]))
return list(edges_in_shortest_paths)Big(O) Analysis
Edge Cases
| Case | How to Handle |
|---|---|
| Null or empty graph (no nodes or edges) | Return an empty list immediately as there are no paths to consider. |
| Source and destination are the same node | If source and destination are the same, any edge connected to that node is on a shortest path of length 0, so return those edges, or empty list if no such edges exist. |
| Graph is disconnected, no path exists between source and destination | Dijkstra's or similar shortest path algorithms will not find a path; return an empty list. |
| Negative edge weights (Dijkstra's algorithm will fail) | Dijkstra's algorithm is not suitable for graphs with negative edge weights; consider using Bellman-Ford, and if a negative cycle is detected, handle appropriately. |
| Graph contains cycles | Dijkstra's and Bellman-Ford algorithms can handle cycles; the shortest path algorithm should not get stuck in a cycle unless it's a negative cycle when using Bellman-Ford. |
| Multiple shortest paths exist between the source and destination using different edges | The algorithm needs to identify and collect all edges that participate in *any* of the shortest paths. |
| Integer overflow when calculating path distances with large weights or many edges | Use a larger data type (e.g., long long in C++) or appropriate overflow handling to prevent incorrect shortest path calculations. |
| Large graph with many nodes and edges (scalability) | The shortest path algorithm should be efficient (e.g., Dijkstra's with a priority queue) to handle large graphs within reasonable time and memory constraints. |